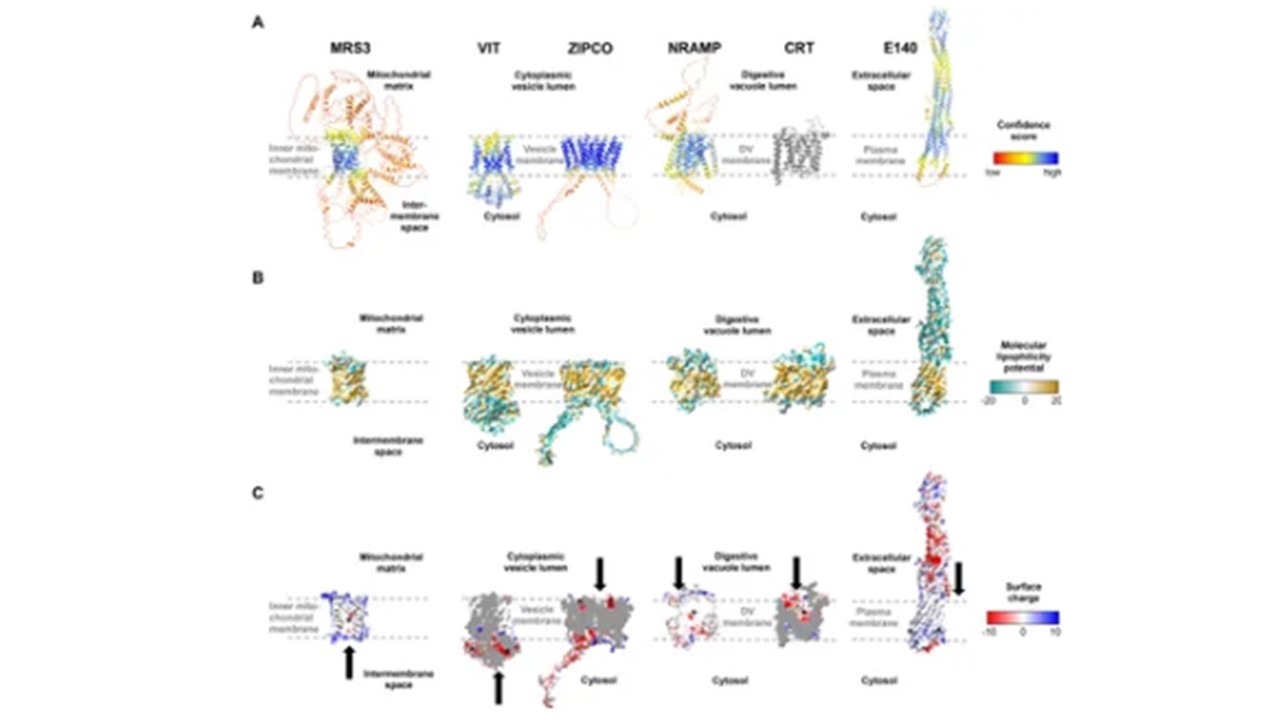
Structures of known and putative P. falciparum iron transporters as viewed from the membrane plane. (A) Predicted protein structures with per-residue pLDDT (predicted local distance difference test) confidence scores on a scale from 0 to 100, where blue represents high and red low confidence, respectively. The experimentally determined structure of PfCRT is shown in gray. (B) Molecular lipophilicity potential of the protein surfaces as implemented in UCSF ChimeraX; tan is hydrophobic and cyan hydrophilic. Dashed lines above and below the tan regions of all proteins indicate the respective membrane and disordered loops were removed for clarity. (C) Surface charge of the proteins with positively charged areas colored blue and negatively charged ones red. Putative cation-binding site are indicated with an asterisk and transport directions by arrows. PfE140 likely forms a dimer but is shown as a monomer, as no predicted dimer structure could be obtained using AlphaFold2-multimer. The putative cation-binding sites for this protein are based on DeepFRI gradCAM scores for the functional term GO:0015075 “monoatomic ion transmembrane transporter activity, Wunderlich J, Kotov V, Votborg-Novél L, Ntalla C, Geffken M, Peine S, Portugal S, Strauss J. Iron transport pathways in the human malaria parasite Plasmodium falciparum revealed by RNA-sequencing. Front Cell Infect Microbiol. 2024 14:1480076. PMID: 39575308
Other associated proteins
| PFID | Formal Annotation |
|---|---|
| PF3D7_0104100 | protein e140, putative |
| PF3D7_0709000 | chloroquine resistance transporter |
| PF3D7_0905200 | mitochondrial carrier protein, putative |
| PF3D7_1022300 | ZIP domain-containing protein, putative, ZIPCO |
| PF3D7_1223700 | vacuolar iron transporter. VIT |