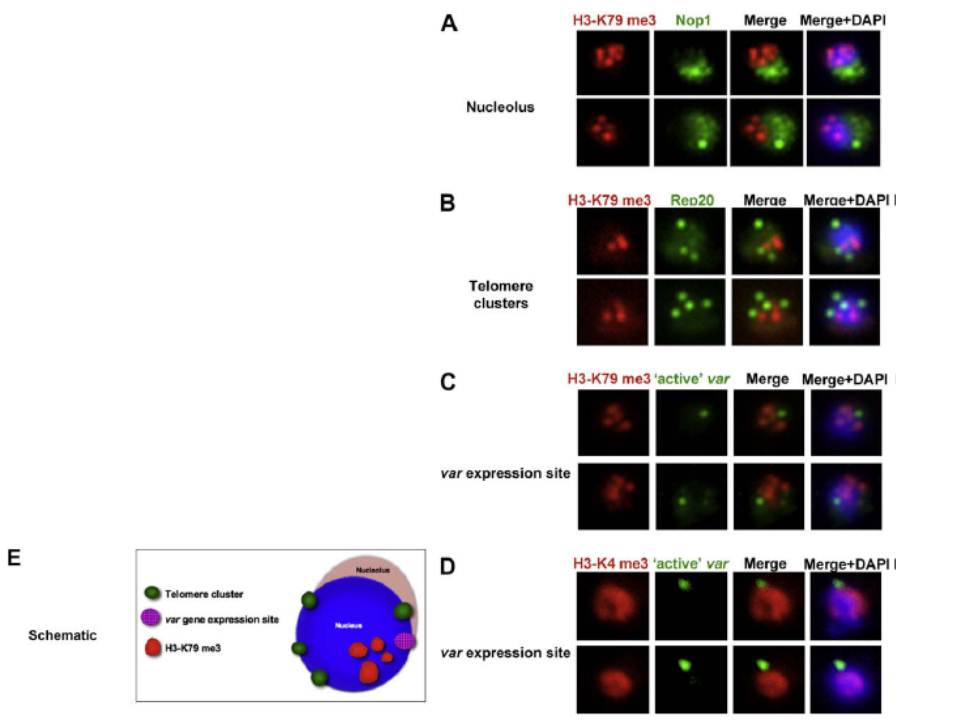
Co-localisation of H3-K79me3 with known cellular compartments. A. Nucleolus: Double labelling IFAs with antibodies against H3-K79me3 and nucleolus marker, PfNop1. B. Telomere clusters: IFeFISH analysis between H3-K79me3 (red) and Rep20 (green). C. Active var gene: IF-FISH analysis between H3-K79me3 (red) and active var gene (green). D. Active var gene colocalisation with activating mark, H3-K4me3. IF-FISH analysis between H3-K4me3 (red) and active var gene (green). DAPI stained nucleus in blue. Font colour represents corresponding fluorescein labelled antibody. Scale bar depicts 1 mm. E. Schematic of the dynamic compartmen-talisation of the P. falciparum nucleus. The distinct sub-nuclear distribution of histone methylation modification, taking example of H3-K79me3 as observed in this study. Known compartments compatible with active transcription are indicated. H3-K79me3 occupies a distinct zone.
Issar N, Ralph SA, Mancio-Silva L, Keeling C, Scherf A. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes Infect. 2009 11:403-7.
Other associated proteins
| PFID | Formal Annotation |
|---|---|
| PF3D7_0617900 | histone H3 variant |
| PF3D7_1105000 | histone H4 |
| PF3D7_1200600 | erythrocyte membrane protein 1, PfEMP1 |
| PF3D7_1333700 | histone H3-like centromeric protein CSE4 |
| PF3D7_1407100 | rRNA 2'-O-methyltransferase fibrillarin, putative |
| Rep20 | Rep20 |