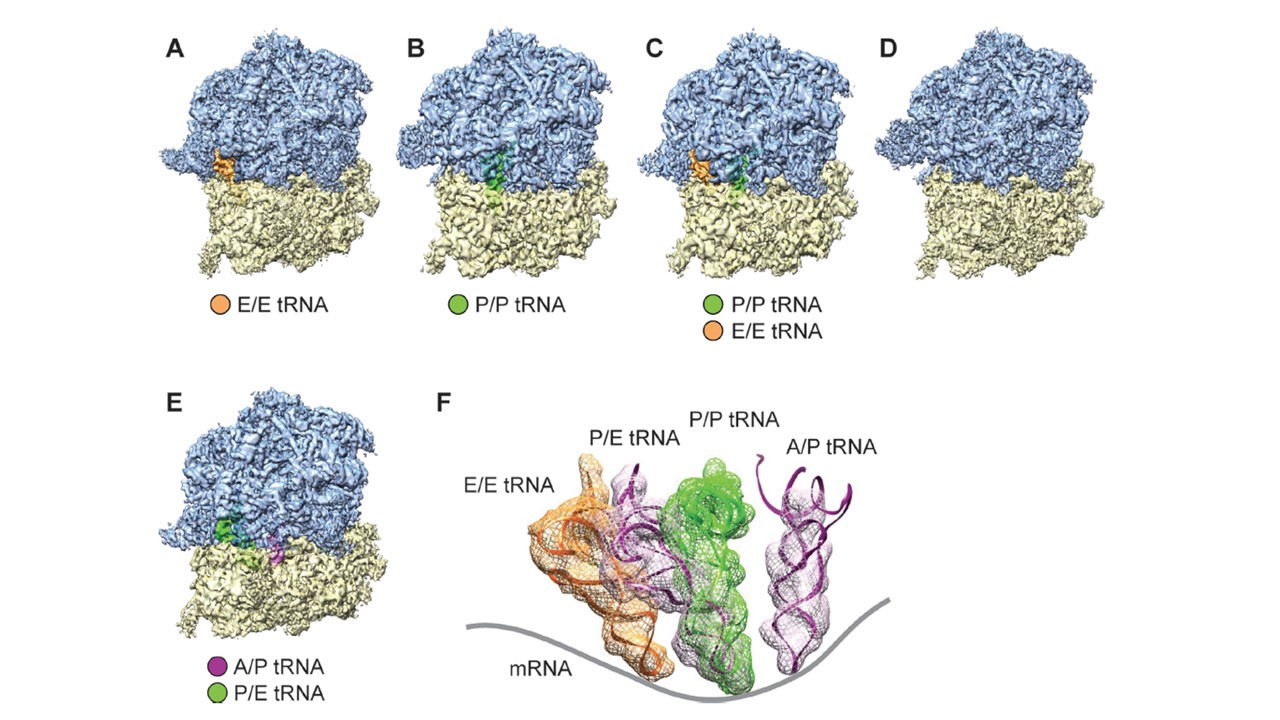
Cryo-EM reconstructions of P. falciparum 80S ribosomes obtained by classification. (A–D) Density maps of the P. falciparum 80S ribosome in non-rotated states (A) bound with E-tRNA (96 732 particles; 53.4%) at an average resolution of 4.7 A˚ ; (B) bound with P-tRNA (14 696 particles; 8.1%) at 6.7 A˚ ; (C) bound with P/P- and E/E-tRNAs (14 676 particles; 8.1%) at 6.7 A˚ ; and (D) without tRNAs (32 246 particles; 17.8%) 5.1 A˚ . (E) rotated state (22 793 particles; 12.6%) at 5.8 ˚A resolution. 60S subunits are colored in blue and 40S subunits are in yellow. (F) Positions of tRNAs for all 80S complexes in (A)–(D). The structures of E-tRNA, P-tRNA and P/E-tRNA were obtained byMDFF fitting, and the structure of A/P-tRNA is from the existing model, PDB 3J0Z, rigid body-fitted into the segmented map in UCSF Chimera. Contours of cryo-EM densities are displayed in mesh; structures of tRNA are displayed as ribbons; and mRNA path has been added as cartoon.
Sun M, Li W, Blomqvist K, Das S, Hashem Y, Dvorin JD, Frank J. Dynamical features of the Plasmodium falciparum ribosome during translation. Nucleic Acids Res. 2015 43(21):10515-24. PMID: 26432834
Other associated proteins
| PFID | Formal Annotation |
|---|---|
| PF3D7_0622800 | leucine--tRNA ligase, putative |
| PF3D7_1331700 | glutamine--tRNA ligase, putative |